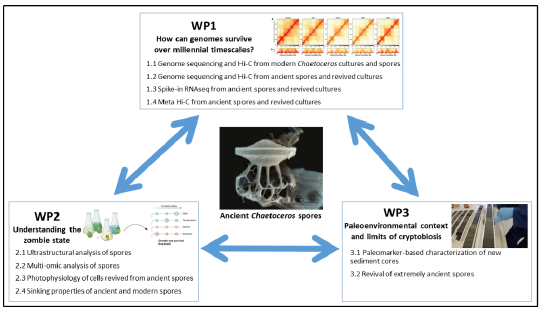
This project aims to investigate the genomic and physiological adaptations of marine diatoms over evolutionary timescales, with a particular focus on Chaetoceros resting spores that we have revived from Antarctic sediment cores spanning the last approx. 10,000 years, and possibly extending further back as far as 3.5 million years. We hypothesize 1) that these spores retain genomic and metabolic signatures that have allowed them to survive extended dormancy, and 2) have retained genotypes tuned to past environmental conditions. Both aspects will be explored. Using both ancient spores and cultures revived from them, we will perform whole genome sequencing, spike-in transcriptomics, and Hi-C to analyse genome structure and gene expression. In parallel we will explore the metabolome and ultrastructure of ancient spores. Phenotypic characteristics of revived cultures, including an assessment of their photophysiologies, will complement these analyses, providing a comprehensive understanding of the adaptive capacities of ancient genotypes to major extrinsic factors such as temperature, sea ice, light, salinity, and nutrient availability. Findings from these analyses will be correlated with past conditions through paleomarker proxies from the sediment cores. The ultimate goal is to link long-term genomic resilience with ecological function, offering insights into the future adaptability of a keystone marine species facing rapid climate change. More generally, we will reveal for the first time how a eukaryotic genome can remain in a viable state for millennia, and aim to extend the limits of organism survival way beyond currently known limits.
ANR DIAZOM
ANR DIAZOM
PALEO, Xavier Crosta